Research in the Tucker-Kellogg lab at Duke-NUS
For pressure ulcer research, we study muscle injury, oxidative stress, and tissue regeneration, using pre-clinical model wounds as well as computational modeling. Pressure ulcers are classified as chronic wounds, and the reasons for poor healing are not well understood. There is an unmed need for strategies to assess risk, improve healing, and save the viable tissue from dying. We welcome collaboration with others, in addition to our self-sufficient agenda of experiments and modeling. Click to learn more.
-
Collaborative Science: Mathematical Modeling of Cancer Plasticity --
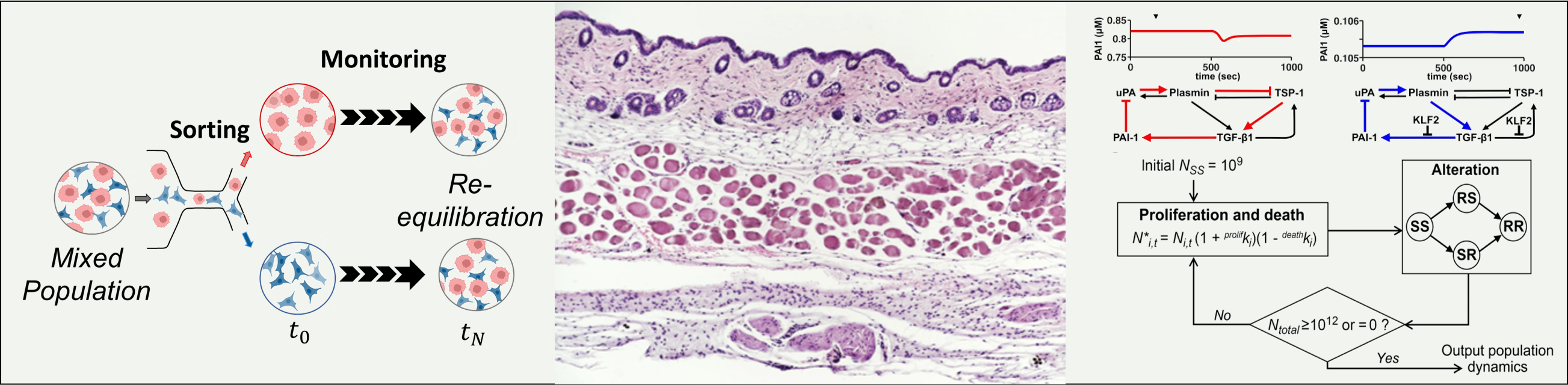
We perform mathematical modeling for a variety of molecular biology applications, with special emphasis on cancer plasticity and biochemical networks. We use methods of population dynamics to study clonal expansion and the evolution of subpopulations of cells that resist drugs or combinations of drugs. We use Markov models to decribe the stochastic transitions of cell state, such as mutations or phenotype changes. We use differential equation models to decribe the timing and magnitude of activation and inhibition relationships in systems biology. We use metabolic flux models to analyze metabolite abundance data for interpreting pathway utilization. Overall, these approaches allow integration of isolated facts and scale-up of cause-and-effect reasoning, to examine different hypothesis for complex cellular behaviors. We collaborate with experimental partners to create back-and-forth between modeling and experiments. Click to learn more.